Page 87 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 87
11.4 Positional Cloning 381
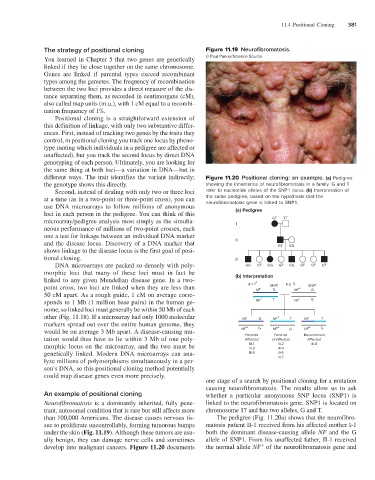
The strategy of positional cloning Figure 11.19 Neurofibromatosis.
You learned in Chapter 5 that two genes are genetically © Paul Parker/Science Source
linked if they lie close together on the same chromosome.
Genes are linked if parental types exceed recombinant
types among the gametes. The frequency of recombination
between the two loci provides a direct measure of the dis-
tance separating them, as recorded in centimorgans (cM),
also called map units (m.u.), with 1 cM equal to a recombi-
nation frequency of 1%.
Positional cloning is a straightforward extension of
this definition of linkage, with only two substantive differ-
ences. First, instead of tracking two genes by the traits they
control, in positional cloning you track one locus by pheno-
type (noting which individuals in a pedigree are affected or
unaffected), but you track the second locus by direct DNA
genotyping of each person. Ultimately, you are looking for
the same thing at both loci—a variation in DNA—but in
different ways. The trait identifies the variant indirectly; Figure 11.20 Positional cloning: an example. (a) Pedigree
the genotype shows this directly. showing the inheritance of neurofibromatosis in a family. G and T
Second, instead of dealing with only two or three loci refer to nucleotide alleles of the SNP1 locus. (b) Interpretation of
at a time (as in a two-point or three-point cross), you can the same pedigree, based on the hypothesis that the
use DNA microarrays to follow millions of anonymous neurofibromatosis gene is linked to SNP1.
loci in each person in the pedigree. You can think of this (a) Pedigree
microarray/pedigree analysis most simply as the simulta- I GT TT
neous performance of millions of two-point crosses, each
one a test for linkage between an individual DNA marker
and the disease locus. Discovery of a DNA marker that II GT GG
shows linkage to the disease locus is the first goal of posi-
tional cloning. III
DNA microarrays are packed so densely with poly- GG GT GG GT GG GT GT GT
morphic loci that many of these loci must in fact be
linked to any given Mendelian disease gene. In a two- (b) Interpretation
point cross, two loci are linked when they are less than II-1 NF SNP1 II-2 NF + SNP1
G
G
50 cM apart. As a rough guide, 1 cM on average corre-
sponds to 1 Mb (1 million base pairs) in the human ge- NF + T NF + G
nome, so linked loci must generally be within 50 Mb of each
other (Fig. 11.18). If a microarray had only 1000 molecular NF G NF + T NF T
markers spread out over the entire human genome, they + + +
G
NF
would be on average 3 Mb apart. A disease-causing mu- NF Parental G NF G Recombinant
Parental
tation would thus have to lie within 3 Mb of one poly- A ected Una ected A ected
III-1
morphic locus on the microarray, and the two must be III-3 III-2 III-8
III-4
genetically linked. Modern DNA microarrays can ana- III-5 III-6
III-7
lyze millions of polymorphisms simultaneously in a per-
son’s DNA, so this positional cloning method potentially
could map disease genes even more precisely.
one stage of a search by positional cloning for a mutation
causing neurofibromatosis. The results allow us to ask
An example of positional cloning whether a particular anonymous SNP locus (SNP1) is
Neurofibromatosis is a dominantly inherited, fully pene- linked to the neurofibromatosis gene. SNP1 is located on
trant, autosomal condition that is rare but still affects more chromosome 17 and has two alleles, G and T.
than 100,000 Americans. The disease causes nervous tis- The pedigree (Fig. 11.20a) shows that the neurofibro-
sue to proliferate uncontrollably, forming tumorous bumps matosis patient II-1 received from his affected mother I-1
under the skin (Fig. 11.19). Although these tumors are usu- both the dominant disease-causing allele NF and the G
ally benign, they can damage nerve cells and sometimes allele of SNP1. From his unaffected father, II-1 received
+
develop into malignant cancers. Figure 11.20 documents the normal allele NF of the neurofibromatosis gene and