Page 89 - Genetics_From_Genes_to_Genomes_6th_FULL_Part3
P. 89
11.4 Positional Cloning 383
Figure 11.21 Some noninformative matings. (a) Even if Obtaining sufficient pedigree data
you know that the NF gene and SNP2 are linked and you know With millions of polymorphic loci on a DNA chip, it should
the phase (arrangement of alleles) in the parents, the mating may
or may not be informative. If the child is a CC or AA homozygote, be possible in theory to map disease genes very accurately,
the cross would be informative. If the child is an AC heterozygote, even if certain crosses are uninformative for linkage of the
you don’t know which allele came from which parent, so the disease gene to certain DNA markers. However, the resolu-
cross would be noninformative. (b) If neither parent is a double tion of positional cloning is always limited in practice by
heterozygote, it is impossible to perform linkage analysis. (SNP3 the number of people human geneticists can track in fami-
is a third SNP locus.) lies in which the disease is segregating. If scientists have
(a)
mapped a disease gene to within 1 cM of a DNA polymor-
II-1 II-2 phism, this value means they have examined the pheno-
SNP2 SNP2
NF C NF + C types (affected or unaffected) of at least 100 members of
such families, and they have also genotyped on microarrays
NF + A NF + A the DNA of all these people. (Remember that 1 cM means
1 recombinant gamete out of 100 total gametes.)
For this reason, positional cloning achieved its first
CC CC AA AA AC AC successes for diseases that can be found in extended
P R R P ? ? families with a large number of children. In 1984, the Hun-
Informative Noninformative tington disease (HD) locus became the first human disease
gene to be mapped successfully by positional cloning pre-
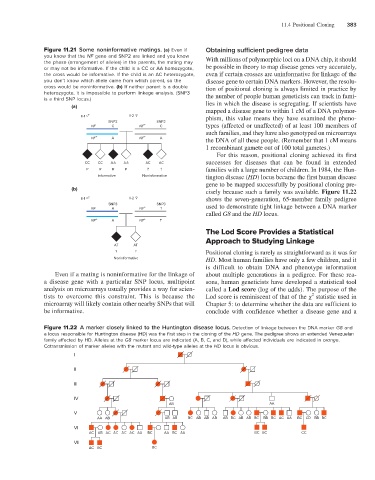
(b) cisely because such a family was available. Figure 11.22
II-1 II-2 shows the seven-generation, 65-member family pedigree
SNP3 SNP3 used to demonstrate tight linkage between a DNA marker
NF A NF + T
called G8 and the HD locus.
NF + A NF + T
The Lod Score Provides a Statistical
Approach to Studying Linkage
AT AT
? ? Positional cloning is rarely as straightforward as it was for
Noninformative HD. Most human families have only a few children, and it
is difficult to obtain DNA and phenotype information
Even if a mating is noninformative for the linkage of about multiple generations in a pedigree. For these rea-
a disease gene with a particular SNP locus, multipoint sons, human geneticists have developed a statistical tool
analysis on microarrays usually provides a way for scien- called a Lod score (log of the odds). The purpose of the
2
tists to overcome this constraint. This is because the Lod score is reminiscent of that of the χ statistic used in
microarray will likely contain other nearby SNPs that will Chapter 5: to determine whether the data are sufficient to
be informative. conclude with confidence whether a disease gene and a
Figure 11.22 A marker closely linked to the Huntington disease locus. Detection of linkage between the DNA marker G8 and
a locus responsible for Huntington disease (HD) was the first step in the cloning of the HD gene. The pedigree shows an extended Venezuelan
family affected by HD. Alleles at the G8 marker locus are indicated (A, B, C, and D), while affected individuals are indicated in orange.
Cotransmission of marker alleles with the mutant and wild-type alleles at the HD locus is obvious.
I
II
III
IV
AB AA
V
AA AB AB AB BC AB AB ABB AB BC ABAB BC BB BC AC AA BC CD BB BC
VI
AC AB AC AC AC AC AA BC AA BC AA BC BC CC
VII
AC BC BC